This is the multi-page printable view of this section. Click here to print.
Instruments
1 - RC MILabsU-CT
The microCT data needed to be transferred to the RDS before uploading from RDS to XNAT. Both XNAT and RDS will keep a copy of microCT data at the current stage. If RDS storage researches its maximum capacity or budget in the future, the microCT data on RDS will be removed periodically to save the space.
The microCT data can be categorized into three types: raw data, temporary data and reconstructed data.
The raw data is saved on the acquisition computer, and in the D drive of the reconstruction computer.
The temporary data is generated in the reconstruction process including the “corr” and “prev” folder of each scan and saved in the “ct-data” folder. “corr” folder contains all the projection correction files, while “prev” folder contains the single slice preview image of reconstruction. “corr” and “prev” folder will not be uploaded to the RDS and XNAT.
The reconstructed data contains the final images for CT reconstruction, and it is saved in the “Results” folder of each scan. Data is uploaded from microCT to RDS daily at 7 p.m.
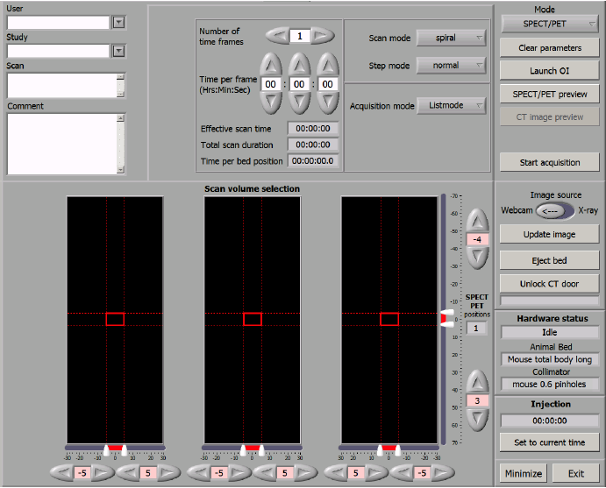
Acquisition control screen
The screenshot above is the acquisition control screen. The User field should always leave blank. The Study field should be filled with the XNAT
Project ID and Subject Name in XNAT, e.g. 123_SubjectName.
Project name can’t contain _, this symbol is used to separate the Project ID and Subject Name. Subject Name can contain any numbers of _.
The Scan name will become the Session name on XNAT.
Instrument data hierarchy is:
Z: (Aquisiton) D: (Reconstruction)
D:\Data\[Study]\[Scan]\*
[Study] is [XNAT Project ID]_[Subject]
[Scan] is [Date]_[Other]
Metadata
| Instrument Source | Logic | Config Source | XNAT Destination |
|---|---|---|---|
[Study] | [XNAT Project ID]_* | - | Subject |
[Study] | *_[Subject] | Subject | |
[Scan] | - | - | Session |
| - | - | TBD | [IID] |
| - | - | TBD | [QCPID] |
[Scan] | [Date]_* | - | Date |
| - | - | - | Time |
[Scan].log | …User: [Operator]… (Not collecting this for now) | - | Operator |
| - | - | TBD | Scanner Name |
| - | - | TBD | Scanner Type |
| - | - | Sydney Imaging | Acquisition Site |
Data
| Original structure | Note | XNAT structure | XNAT File Type | |
|---|---|---|---|---|
[Scan].ct-sequence | [Scan].zip | uct | RAW | |
[Scan].zipuct | [Scan].zip | uct | RAW | |
[Scan].mCT | [Scan].zip | uct | RAW | |
[Scan].parameters | [Scan].zip | uct | RAW | |
[Scan].log | [Scan].zip | uct | RAW | |
[Scan].comments | [Scan].zip | uct | RAW | |
[Scan]_left.png | [Scan].zip | uct | RAW | |
[Scan]_right.png | [Scan].zip | uct | RAW | |
[Scan]_top.png | [Scan].zip | uct | RAW | |
[Scan]_x-ray-left.png | [Scan].zip | uct | RAW | |
[Scan]_x-ray-right.png | [Scan].zip | uct | RAW | |
[Scan]_x-ray-top.png | [Scan].zip | uct | RAW | |
ct-data/calibration/[Stuff].smv | calibration | |||
ct-data/calibration/[stuff2]_dark.tif | calibration | |||
ct-data/calibration/[stuff2]_white.tif | calibration | |||
ct-data/calibration/geopar.cfg | calibration | |||
ct-data/proj_000_0_log.csv | proj | |||
ct-data/proj_000_0_00_[NNN].tif | where NNN is the index | proj | ||
Results/CT_[Scan]_80um.log | Not maintaining folder structure | CT_[Scan]_80um.log | log | RECON |
Results/CT_[Scan]_80um.nii | Not maintaining folder structure | CT_[Scan]_80um.nii | NIFTI | RECON |
2 - MR Solutions 3T & 7T MRI
[Series ID] should be used for matching files.
The Project field should filled with the XNAT shortcode (EZBooking Project ID) and the Subject field will be filled with the project name.
Note
For the new project created through DashR, the XNAT shortcode should be at least 3 digits/characters and less than 20 digits/characters.
If less than 3 digits, zeros should be added at the beginning. For example, XNAT shortcode 044 for EZBooking Project ID 44
Metadata
| Instrument Source | Logic | Config Source | XNAT Destination |
|---|---|---|---|
| (0010,0010) | [Project]:* | - | [Project] |
| (0010,0010) | *:[Subject] | - | [Subject] |
| (0008,0020) | [YYYYMMDD] | [Session] | |
| (0008,0050) | [Subject Accession Number] | - | |
| - | - | 3T: 10.25910/5cf9e65ffa8c4 7T: 10.25910/5cf9f821b4c94 | [IID] TBD |
| - | - | TBD | [QCPID] TBD |
| TBC | - | - | Date |
| Timestamp (0008,0032) TBC | - | - | Time |
| Acquisition Duration (0018,9073) | - | - | TBD |
| Protocol Name (0018,1030) /Sequence Name (0018,0024) | - | - | TBD |
| - | - | - | Operator |
| (0008,1010) Station Name | - | - | Scanner Name |
| (0008,1090) Manufacturer’s Model Name | - | - | Scanner Type |
| - | - | Sydney Imaging | Acquisition Site |
Data
| Original structure | Note | XNAT structure | XNAT File Type |
|---|---|---|---|
D:\smis\dev\Data\[Subject ID]\DICOM\[Series]\[1]\[Series]_[ScanX] | DICOM | \Session\[Series]\ | DICOM |
D:\smis\dev\Data\[Subject ID]\Image\[Series]\[1]\[Series]_[ScanX] | SUR | \Session\[Series]\ | SUR |
D:\smis\dev\Data\[Subject ID]\Image\[Series]\patient[SubjectID]_scan[Series] | PET (log) | ||
D:\smis\dev\Data\[Subject ID]\Image\[Series]\[Reconstruction]\[Series]_stuff | PET (Reconstructed) | ||
D:\smis\dev\Data\[Subject ID]\Spectroscopy\[Series]\[Series]_0 | Spectroscopy | \Session\[Series]\ | MRD, SPR |
D:\smis\dev\MRD\[Subject ID]\Image\[Series]\[Series]_0 | MRD, RPR, rtv | \Session\[Series]\ | MRD, RPR |
D:\smis\dev\MRD\[Subject ID]\Image\[Series]\[Series]_0_o | MRD, RPR, rtv | \Session\[Series]\ | MRD, RPR |
D:\smis\dev\MRD\[Subject ID]\Image\[Series]\btable.txt | MRD, RPR, rtv | \Session\[Series]\ | btable |
D:\smis\dev\MRD\[Subject ID]\Image\[Series]\rtable.txt | MRD, RPR, rtv | \Session\[Series]\ | rtable |
D:\smis\dev\PetRaw\[Subject ID]\[Series]\pet_[Series] | PET (RAW) |
3 - Siemens ARTIS pheno
Metadata
| Instrument Source | Logic | Config Source | XNAT Destination |
|---|---|---|---|
| Additional Info field: Project:HT-003 | (0010,4000) Patient’s Comments | (0010,4000) Patient’s Comments | Project: HT-003 |
| Lastname:Mukherjee Firstname:Sheep1 Title: | (0010,0010) Patient’s Name | (0010,0010) Patient’s Name | Subject: Mukherjee_sheep1 |
| Patient ID: Patient01 | (0010,0020) Patient ID | (0010,0020) Patient ID + (0032,0012) Study ID Issuer | Session: Patient01_210301-15_47_19-DST-IVS |
| (0020,0010) Study UID | (0020,0010) Study UID | SCAN ID: 990 | |
| 10.25910/5cf9f821b4c94 | [IID] TBD | ||
[QCPID] TBD |
[Path] needs to be determined. It can be on instrument, or the export location on RDS.
Data
| Original structure | Note | XNAT structure |
|---|---|---|
[Path]//[File].jpg | To be covered with DICOM2JPEG | Resource file |
[Path]//[File].avi | Resource file | |
[Path]//[File].txt | Resource file | |
[Path]//[File].csv | Resource file |